Extract-Transform-Load (ETL) with Python
Read, Clean, Create Schema and Load bunch of csv files to MySQL with Python
I wanted to create a staging area in mysql to build Datawarehouse from bunch of csv files.
Before uploading the data in mysql I would want to perform some data quality check.
First Import following libraries
import numpy as np
import pandas as pd
## set the connection to the db
import sqlalchemy
import pymysql
from IPython.display import Image
Since CMS released the claims payment information for 2012, CMS has received a growing number of data requests for provider enrollment data, and there is a growing interest from the health care industry to identify Medicare-enrolled providers and suppliers and their associations to groups/organizations.
CMS continues to move toward data transparency for all non-sensitive Medicare provider and supplier enrollment data. This aligns with the agency’s effort to promote and practice data transparency for Medicare information. Publishing this data allows users, including other health plans, to easily access and validate provider information against Medicare data.
The Public Provider Enrollment files will include enrollment information for providers and suppliers who were approved to bill Medicare at the time the file was created. The data elements on the files are disclosable to the public. This data will focus on data relationships as they relate to Medicare provider enrollment.
The provider enrollment data will be published on https://data.cms.gov/public-provider-enrollment and will be updated on a quarterly basis. The initial data will consist of individual and organization provider and supplier enrollment information similar to what is on Physician Compare; however, it will be directly from PECOS and will only be updated through updates to enrollment information. Elements will include:
lets import a example csv file
example_df = pd.read_csv('Data/Clinic_Group_Practice_Reassignment_A-D.csv')
example_df.head(6)
| Group PAC ID | Group Enrollment ID | Group Legal_Business Name | Group State Code | Group Due Date | Group Reassignments and Physician Assistants | Record Type | Individual Enrollment ID | Individual NPI | Individual First Name | Individual Last Name | Individual State Code | Individual Specialty Description | Individual Due Date | Individual Total Employer Associations | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1153364203 | O20050609000145 | A & A Audiology, Pc | TX | NaN | 1 | Reassignment | I20050609000172 | 1.306861e+09 | Tonia | Fleming | TX | Audiologist | NaN | 2 |
| 1 | 1759440886 | O20081105000006 | A & A Chiropractic. Llc | NJ | NaN | 1 | Reassignment | I20080819000132 | 1.134400e+09 | Sharon | Barnum | NJ | Chiropractic | NaN | 1 |
| 2 | 1153442256 | O20101222000765 | A & A Eye Associates, Pc | PA | 09/30/2017 | 2 | Reassignment | I20101222000889 | 1.538155e+09 | Daniel | Anderson | PA | Optometry | 09/30/2017 | 1 |
| 3 | 1153442256 | O20101222000765 | A & A Eye Associates, Pc | PA | 09/30/2017 | 2 | Reassignment | I20091118000149 | 1.407852e+09 | Amanda | Temnykh | PA | Optometry | 07/31/2016 | 1 |
| 4 | 3274425004 | O20040326000876 | A & A Health Systems, Inc | MS | NaN | 1 | Reassignment | I20040329000500 | 1.063480e+09 | Sheryll | Vincent | MS | Pediatric Medicine | NaN | 1 |
| 5 | 4082882519 | O20110727000379 | A & A Hearing Group, Ps | WA | 06/30/2016 | 2 | Reassignment | I20110727000462 | 1.396781e+09 | Ashley | Al Izzi | WA | Audiologist | 08/31/2016 | 1 |
Lets look at the data types of dataframe
dtype_pd = pd.DataFrame(example_df.dtypes, columns = ['data_type']).reset_index()
unique_records = pd.DataFrame(example_df.nunique(), columns = ['unique_records']).reset_index()
info_df = pd.merge(dtype_pd, unique_records, on = 'index')
info_df
| index | data_type | unique_records | |
|---|---|---|---|
| 0 | Group PAC ID | int64 | 59730 |
| 1 | Group Enrollment ID | object | 63038 |
| 2 | Group Legal_Business Name | object | 59176 |
| 3 | Group State Code | object | 55 |
| 4 | Group Due Date | object | 35 |
| 5 | Group Reassignments and Physician Assistants | object | 542 |
| 6 | Record Type | object | 2 |
| 7 | Individual Enrollment ID | object | 450876 |
| 8 | Individual NPI | float64 | 429093 |
| 9 | Individual First Name | object | 39181 |
| 10 | Individual Last Name | object | 134023 |
| 11 | Individual State Code | object | 56 |
| 12 | Individual Specialty Description | object | 169 |
| 13 | Individual Due Date | object | 35 |
| 14 | Individual Total Employer Associations | int64 | 51 |
It seams that there’s no primary key in our DataFrame lets check that:
print('Is there a column with unique values in entire dataFrame? : ', len(example_df) in info_df['unique_records'])
Is there a column with unique values in entire dataFrame? : False
example_df.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| Group PAC ID | 552790.0 | 4.962019e+09 | 2.888691e+09 | 4.210108e+07 | 2.567352e+09 | 4.880689e+09 | 7.517196e+09 | 9.931498e+09 |
| Individual NPI | 552788.0 | 1.499834e+09 | 2.881970e+08 | 1.003000e+09 | 1.245784e+09 | 1.497990e+09 | 1.750322e+09 | 1.993000e+09 |
| Individual Total Employer Associations | 552790.0 | 2.927397e+00 | 5.034805e+00 | 1.000000e+00 | 1.000000e+00 | 2.000000e+00 | 3.000000e+00 | 7.800000e+01 |
#Are there duplicate Rows?
print('Total rows repeating is/are', sum(example_df.duplicated()))
Total rows repeating is/are 0
We’ll have to do follow following procedure
#engine = sqlchemy.create_engine('mysql+pymsql://<username>:<password>@<server-name>:<port_number>/<database_name>')
engine = sqlalchemy.create_engine('mysql+pymysql://root:info7370@localhost:3306/try_python')
Image(filename='empty.JPG')
sql_table_name= 'provider'
initial_sql = "CREATE TABLE IF NOT EXISTS " +str(sql_table_name)+ "(key_pk INT AUTO_INCREMENT PRIMARY KEY"
def rename_df_cols(df):
'''Input a dataframe, outputs same dataframe with No Space in column names'''
col_no_space = dict((i, i.replace(' ','')) for i in list(df.columns))
df.rename(columns= col_no_space, index= str, inplace= True)
return df
def dtype_mapping():
'''Returns a dict to refer correct data type for mysql'''
return {'object' : 'TEXT',
'int64' : 'BIGINT',
'float64' : 'FLOAT',
'datetime64' : 'DATETIME',
'bool' : 'TINYINT',
'category' : 'TEXT',
'timedelta[ns]' : 'TEXT'}
#engine = sqlchemy.create_engine('mysql+pymsql://<username>:<password>@<server-name>:<port_number>/<database_name>')
def create_sql(engine, df, sql = initial_sql):
'''input engine: engine (connection for mysql), df: dataframe that you would like to create a schema for,
outputs Mysql schema creation'''
df = rename_df_cols(df)
col_list_dtype = [(i, str(df[i].dtype)) for i in list(df.columns)]
map_data= dtype_mapping()
for i in col_list_dtype:
key = str(df[i[0]].dtypes)
sql += ", " + str(i[0])+ ' '+ map_data[key]
sql= sql + str(')')
print('\n', sql, '\n')
try:
conn = engine.raw_connection()
except ValueError:
print('You have connection problem with Mysql, check engine parameters')
cur = conn.cursor()
try:
cur.execute(sql)
except ValueError:
print("Ohh Damn it couldn't create schema, check Sql again")
cur.close()
create_sql(engine, df= example_df)
CREATE TABLE IF NOT EXISTS provider(key_pk INT AUTO_INCREMENT PRIMARY KEY, GroupPACID BIGINT, GroupEnrollmentID TEXT, GroupLegal_BusinessName TEXT, GroupStateCode TEXT, GroupDueDate TEXT, GroupReassignmentsandPhysicianAssistants TEXT, RecordType TEXT, IndividualEnrollmentID TEXT, IndividualNPI FLOAT, IndividualFirstName TEXT, IndividualLastName TEXT, IndividualStateCode TEXT, IndividualSpecialtyDescription TEXT, IndividualDueDate TEXT, IndividualTotalEmployerAssociations BIGINT)
Image(filename='Schema.JPG')
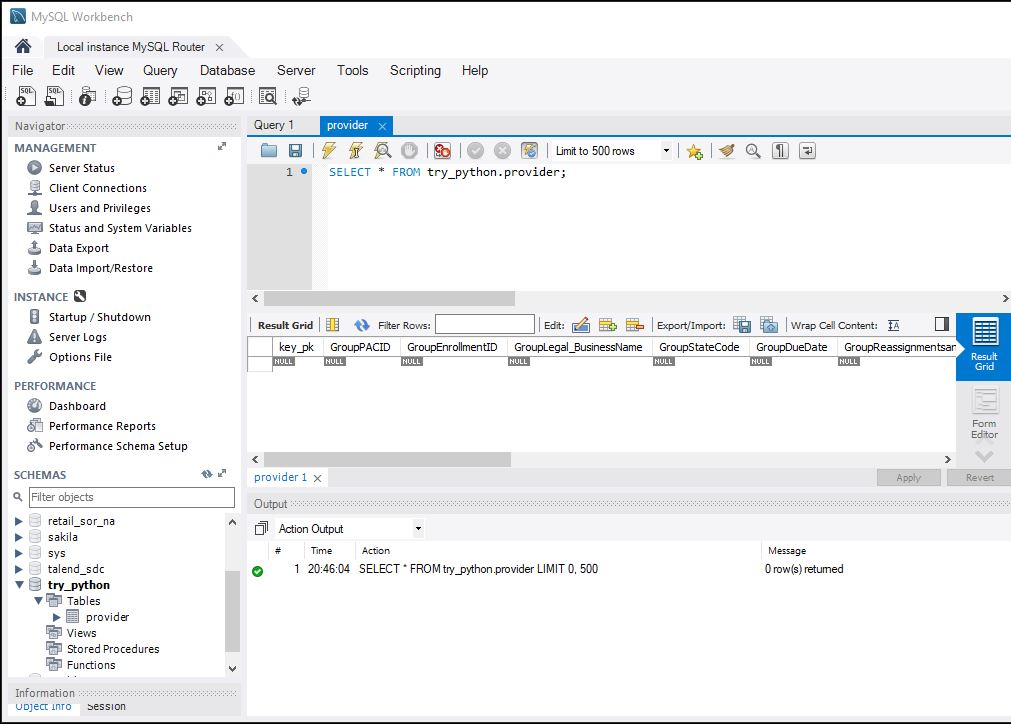
We’ll now load the data in the schema that we just created
import os
from pathlib import Path
mypath = Path().absolute()
data = str(mypath) + str('\\Data\\')
list_of_files = os.listdir(data)
dir_data = [str(data)+str(i) for i in list_of_files if "Clinic_Group_Practice_Reassignment" in i]
def load_data_mysql(dir_data):
for i in dir_data:
i = pd.read_csv(i, low_memory= False)
rename_df_cols(i)
i.to_sql(name= sql_table_name,con = engine, index =False, if_exists = 'append',)
lst = list(i)
del lst
Lets calculate the time to run and later we’ll compare it with SSIS
Image(filename='Loaded.JPG')
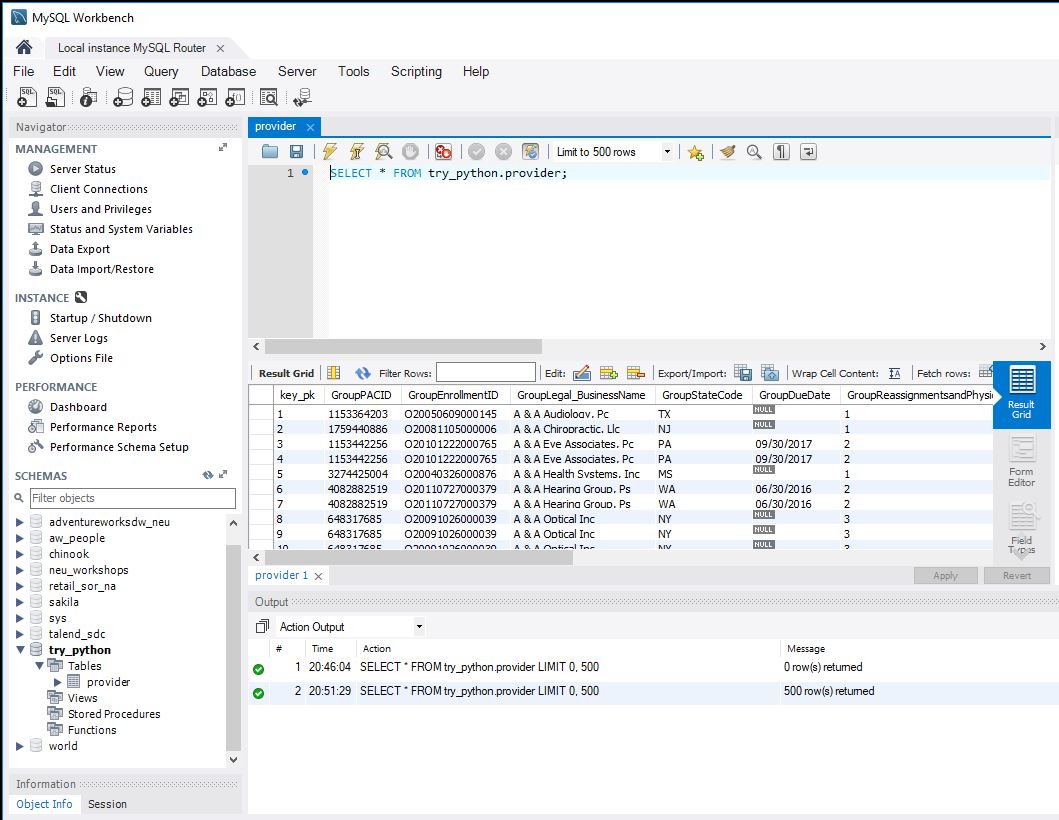
import time
start_time = time.time()
load_data_mysql(dir_data=dir_data)
print("--- %s seconds ---" % (time.time() - start_time))
— 133.69091987609863 seconds —
Now if we try to do the same task using Microsoft SSIS Package. We’ll need to follow these steps:
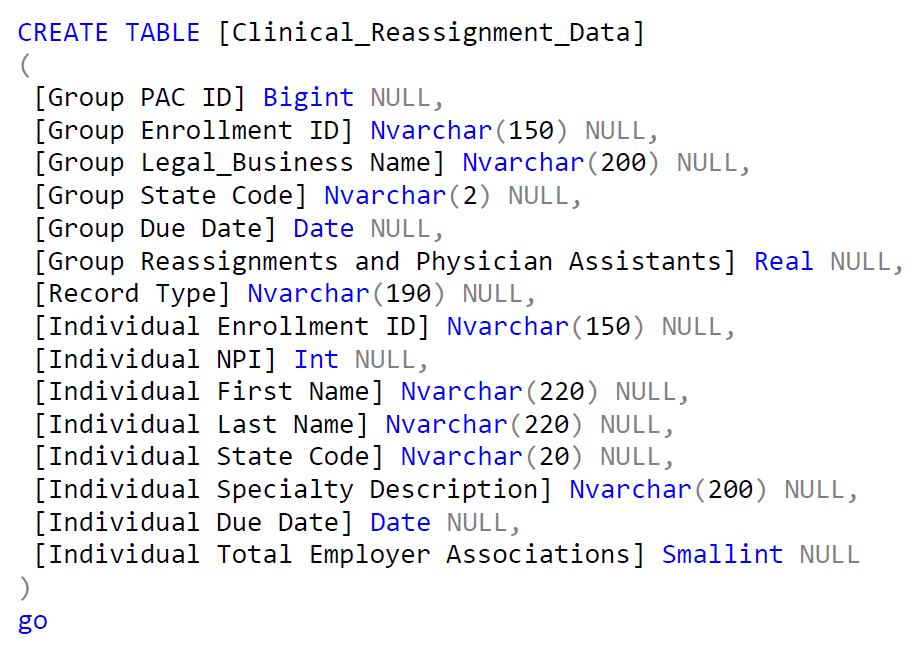
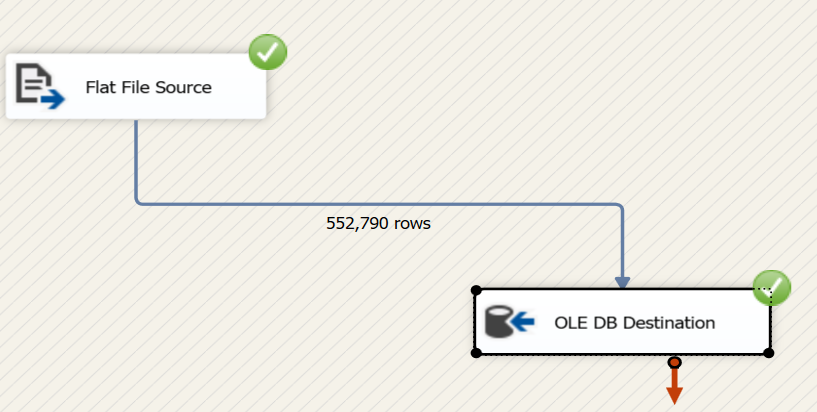
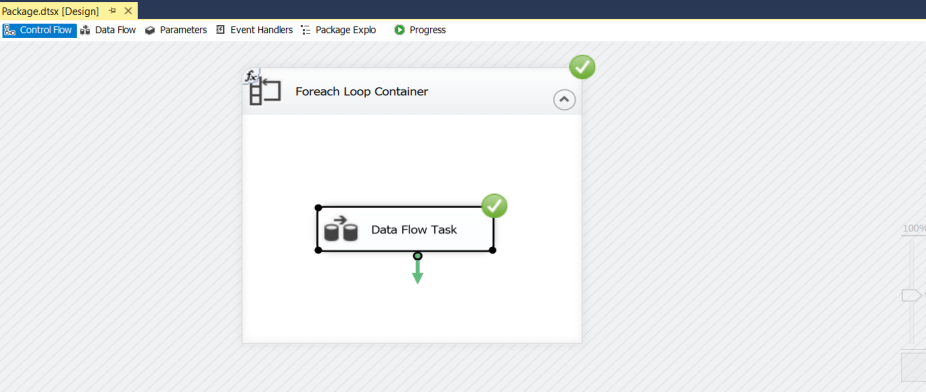
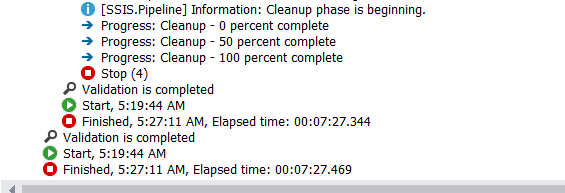
Woah!!
There’s a significant difference in the performance.
With python it took 133 seconds ~ 2 minutes to read, clean and upload, whereas, in SSIS it took 8 minutes.
Python is 400% faster here, with reusable code and completely automated. That’s why I love Python.
— This post ends here—
Read, Clean, Create Schema and Load bunch of csv files to MySQL with Python
Implement Machine learning models to predict interest rates of Bonds from scratch